Abstract
Meningococcal disease remains an important cause of morbidity and death worldwide despite the development and increasing implementation of effective vaccines. Elimination of the disease is hampered by the enormous diversity and antigenic variability of the causative agent, Neisseria meningitidis, one of the most variable bacteria in nature. These features are attained mainly through high rates of horizontal gene transfer and alteration of protein expression through phase variation. The recent availability of whole-genome sequencing (WGS) of large-scale collections of N. meningitidis isolates from various origins, databases to facilitate storage and sharing of WGS data and the concomitant development of effective bioinformatics tools have led to a much more thorough understanding of the diversity of the species, its evolution and population structure and how virulent traits may emerge. Implementation of WGS is already contributing to enhanced epidemiological surveillance and is essential to ascertain the impact of vaccination strategies. This Review summarizes the recent advances provided by WGS studies in our understanding of the biology of N. meningitidis and the epidemiology of meningococcal disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Christensen, H., May, M., Bowen, L., Hickman, M. & Trotter, C. L. Meningococcal carriage by age: a systematic review and meta-analysis. Lancet Infect. Dis. 10, 853–861 (2010).
Cimolai, N. & Caugant D. A. in Laboratory Diagnosis of Bacterial Infections (ed. Cimolai N.) 499–525 (Marcel Dekker, 2001).
Gotschlich, E. C., Goldschneider, I. & Artenstein, M. S. Human immunity to the meningococcus. IV. Immunogenicity of group A and group C meningococcal polysaccharides in human volunteers. J. Exp. Med. 129, 1367–1384 (1969).
Artenstein, M. S. et al. Prevention of meningococcal disease by group C polysaccharide vaccines. N. Engl. J. Med. 282, 417–420 (1970).
Jafri, R. Z. et al. Global epidemiology of invasive meningococcal disease. Popul. Health Metr. 11, 17 (2013).
Wang, B., Santoreneos, R., Giles, L., Haji Ali Afzali, H. & Marshall, H. Case fatality rates of invasive meningococcal disease by serogroup and age: a systematic review and meta-analysis. Vaccine 37, 2768–2782 (2019).
Stephens, D. S., Greenwood, B. & Brandtzaeg, P. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369, 2196–2210 (2007).
Vyse, A., Anonychuk, A., Jäkel, A., Wieffer, H. & Nadel, S. The burden and impact of severe and long-term sequelae of meningococcal disease. Expert Rev. Anti Infect. Ther. 11, 597–604 (2013).
Jyssum, K. & Jyssum, S. Variation in density and transformation potential in deoxyribonucleic acid from Neisseria meningitidis. J. Bacteriol. 90, 1513–1519 (1965).
Feil, E. J., Maiden, M. C., Achtman, M. & Spratt, B. G. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16, 1496–1502 (1999).
Vogel, U. & Frosch, M. The genus Neisseria: population structure, genome plasticity, and evolution of pathogenicity. Curr. Top. Microbiol. Immunol. 264, 23–45 (2002).
Liu, G., Tang, C. M. & Exley, R. M. Non-pathogenic Neisseria: members of an abundant, multi-habitat, diverse genus. Microbiology 161, 1297–1312 (2015).
Linz, B., Schenker, M., Zhu, P. & Achtman, M. Frequent interspecific genetic exchange between commensal Neisseriae and Neisseria meningitidis. Mol. Microbiol. 36, 1049–1058 (2000).
Lu, Q. F. et al. Genus-wide comparative genomics analysis of Neisseria to identify new genes associated with pathogenicity and niche adaptation of Neisseria pathogens. Int. J. Genomics 2019, 6015730 (2019).
Bennett, J. S., Watkins, E. R., Jolley, K. A., Harrison, O. B. & Maiden, M. C. Identifying Neisseria species by use of the 50S ribosomal protein L6 (rplF) gene. J. Clin. Microbiol. 52, 1375–1381 (2014).
Maiden, M. C. & Harrison, O. B. Population and functional genomics of Neisseria revealed with gene-by-gene approaches. J. Clin. Microbiol. 54, 1949–1955 (2016).
Weyrich, L. S. et al. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature 544, 357–361 (2017).
Eerkens, J. W. et al. A probable prehistoric case of meningococcal disease from San Francisco Bay: next generation sequencing of Neisseria meningitidis from dental calculus and osteological evidence. Int. J. Paleopathol. 22, 173–180 (2018).
Schoen, C. et al. Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc. Natl Acad. Sci. USA 105, 3473–3478 (2008).
Schielke, S., Frosch, M. & Kurzai, O. Virulence determinants involved in differential host niche adaptation of Neisseria meningitidis and Neisseria gonorrhoeae. Med. Microbiol. Immunol. 199, 185–196 (2010).
Clemence, M. E. A., Maiden, M. C. J. & Harrison, O. B. Characterization of capsule genes in non-pathogenic Neisseria species. Microb. Genom. 4, e0.000208 (2018).
Lapeysonnie, L. La méningite cérébrospinale en Afrique. Bull. World Health Organ. 28, 1–114 (1963).
Greenwood, B. Manson Lecture. Meningococcal meningitis in Africa. Trans. R. Soc. Trop. Med. Hyg. 93, 341–353 (1999).
Stephens, D. S. Conquering the meningococcus. FEMS Microbiol. Rev. 31, 3–14 (2007).
Harrison, O. B. et al. Description and nomenclature of Neisseria meningitidis capsule locus. Emerg. Infect. Dis. 19, 566–573 (2013).
Tzeng, Y. L. & Stephens, D. S. Epidemiology and pathogenesis of Neisseria meningitidis. Microbes Infect. 2, 687–700 (2000).
Diomandé, F. V. Public health impact after the introduction of PsA-TT: the first 4 years. Clin. Infect. Dis. 61, S467–S472 (2015).
Borrow, R. et al. The Global Meningococcal Initiative: global epidemiology, the impact of vaccines on meningococcal disease and the importance of herd protection. Expert Rev. Vaccines 16, 313–328 (2017).
Krauland, M. G. et al. Whole genome sequencing to investigate the emergence of clonal complex 23 Neisseria meningitidis serogroup Y disease in the United States. PLOS ONE 7, e35699 (2012).
Bröker, M. et al. Meningococcal serogroup Y disease in Europe: continuation of high importance in some European regions in 2013. Hum. Vaccin. Immunother. 11, 2281–2286 (2015).
Mayer, L. W. et al. Outbreak of W135 meningococcal disease in 2000: not emergence of a new W135 strain but clonal expansion within the electophoretic type-37 complex. J. Infect. Dis. 185, 1596–1605 (2002).
Koumaré, B. et al. The first large epidemic of meningococcal disease caused by serogroup W135, Burkina Faso, 2002. Vaccine 25, A37–A41 (2007).
Abad, R., López, E. L., Debbag, R. & Vázquez, J. A. Serogroup W meningococcal disease: global spread and current affect on the Southern Cone in Latin America. Epidemiol. Infect. 142, 2461–2470 (2014).
Krone, M. et al. Increase of invasive meningococcal serogroup W disease in Europe, 2013 to 2017. Euro Surveill. 24, e2807 (2019).
Caugant, D. A. et al. Intercontinental spread of a genetically distinctive complex of clones of Neisseria meningitidis causing epidemic disease. Proc. Natl Acad. Sci. USA 83, 4927–4931 (1986).
Olyhoek, T., Crowe, B. A. & Achtman, M. Clonal population structure of Neisseria meningitidis serogroup A isolated from epidemics and pandemics between 1915 and 1983. Rev. Infect. Dis. 9, 665–692 (1987).
Maiden, M. C. et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl Acad. Sci. USA 95, 3140–3145 (1998).
Yazdankhah, S. P. et al. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J. Clin. Microbiol. 42, 5146–5153 (2004).
Golparian, D. & Unemo, M. Will genome analysis elucidate evolution, global transmission and virulence of Neisseria meningitidis lineages? EBioMedicine 2, 186–187 (2015).
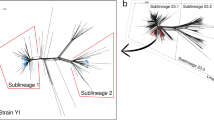
Lucidarme, J. et al. Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J. Infect. 71, 544–552 (2015). This study demonstrates the power of whole genome sequencing to elucidate the global epidemiology of lineage 11.
Bratcher, H. B., Corton, C., Jolley, K. A., Parkhill, J. & Maiden, M. C. A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genom. 15, 1138 (2014).
Topaz, N. et al. Phylogenetic relationships and regional spread of meningococcal strains in the meningitis belt, 2011–2016. EBioMedicine 41, 488–496 (2019).
Jelfs, J., Munro, R., Ashton, F. E. & Caugant, D. A. Genetic characterization of a new variant within the ET-37 complex of Neisseria meningitidis associated with outbreaks in various parts of the world. Epidemiol. Infect. 125, 285–298 (2000).
Ramsay, M. E., Andrews, N., Kaczmarski, E. B. & Miller, E. Efficacy of meningococcal serogroup C conjugate vaccine in teenagers and toddlers in England. Lancet 357, 195–196 (2001).
Retchless, A. C. et al. The establishment and diversification of epidemic-associated serogroup W meningococcus in the African meningitis belt, 1994–2012. mSphere 1, e00201–e00216 (2016).
Mustapha, M. M. et al. Genomic epidemiology of hypervirulent serogroup W, ST-11 Neisseria meningitidis. EBioMedicine 2, 1447–1455 (2015).
Mowlaboccus, S. et al. Clonal expansion of a new penicillin-resistant clade of Neisseria meningitidis serogroup W clonal complex 11, Australia. Emerg. Infect. Dis. 23, 1364–1367 (2017).
Lucidarme, J. An international invasive meningococcal disease outbreak due to a novel and rapidly expanding serogroup W strain, Scotland and Sweden, July to August 2015. Euro Surveill. 21, 30395 (2016).
Eriksson, L. et al. Whole-genome sequencing of emerging invasive Neisseria meningitidis serogroup W in Sweden. J. Clin. Microbiol. 56, e01409–e01417 (2018).
Taha, M.-K., Deghmane, A. E., Knol, M. & van der Ende, A. Whole genome sequencing reveals Trans-European spread of an epidemic Neisseria meningitidis serogroup W clone. Clin. Microbiol. Infect. 25, 765–767 (2019).
Campbell, H., et al. Presentation with gastrointestinal symptoms and high case fatality associated with group W meningococcal disease (MenW) in teenagers, England, July 2015 to January 2016. Euro Surveill. 21, 30175 (2016).
Tzeng, Y. L. et al. Emergence of a new Neisseria meningitidis clonal complex 11 lineage 11.2 clade as an effective urogenital pathogen. Proc. Natl Acad. Sci. USA 114, 4237–4242 (2017). An emerging strain of N. meningitidis causing urethritis in the United States adaptated to the urogenital environment by loss of the capsule and acquisition of the N. gonorrheae norB–aniA cassette promoting anaerobic growth.
Kahler, C. M. Emergence of a urogenital pathotype of Neisseria meningitidis. Trends Microbiol. 25, 510–512 (2017).
Retchless, A. C. et al. Expansion of a urethritis-associated Neisseria meningitidis clade in the United States with concurrent acquisition of N. gonorrhoeae alleles. BMC Genom. 19, 176 (2018).
Moore, P. S., Reeves, M. W., Schwartz, B., Gellin, B. G. & Broome, C. V. Intercontinental spread of an epidemic group A Neisseria meningitidis strain. Lancet 2, 260–263 (1989).
Morelli, G. et al. Clonal descent and microevolution of Neisseria meningitidis during 30 years of epidemic spread. Mol. Microbiol. 25, 1047–1064 (1997).
Caugant, D. A. et al. Molecular characterization of invasive meningococcal isolates from countries in the African meningitis belt before introduction of a serogroup A conjugate vaccine. PLOS ONE 7, e46019 (2012).
Lamelas, A. et al. Emergence of a new epidemic Neisseria meningitidis serogroup A clone in the African meningitis belt: high-resolution picture of genomic changes that mediate immune evasion. mBio 5, e01974-14 (2014). A genomic study showing the evolutionary changes occurring during clonal replacement within a serogroup A lineage in the African meningitis belt and highlighting the role of protein glycosylation for immune evasion.
Watkins, E. R. & Maiden, M. C. Metabolic shift in the emergence of hyperinvasive pandemic meningococcal lineages. Sci. Rep. 7, 41126 (2017).
Diermayer, M. et al. Epidemic serogroup B meningococcal disease in Oregon: the evolving epidemiology of the ET-5 strain. JAMA 281, 1493–1497 (1999).
Rouaud, P. et al. Prolonged outbreak of B meningococcal disease in the Seine-Maritime department, France, January 2003 to June 2005. Euro Surveill. 11, 178–181 (2006).
Harrison, O. B., Bray, J. E., Maiden, M. C. & Caugant, D. A. Genomic analysis of the evolution and global spread of hyper-invasive meningococcal lineage 5. EBioMedicine 2, 234–243 (2015).
Hill, D. M. et al. Genomic epidemiology of age-associated meningococcal lineages in national surveillance: an observational cohort study. Lancet Infect. Dis. 15, 1420–1428 (2015).
Bratcher, H. B. et al. Establishment of the European meningococcal strain collection genome library (EMSC-GL) for the 2011 to 2012 epidemiological year. Euro Surveill. 23 (2018).
Brehony, C. et al. Distribution of Bexsero® antigen sequence types (BASTs) in invasive meningococcal disease isolates: implications for immunisation. Vaccine 34, 4690–4697 (2016).
Yazdankhah, S. P. & Caugant, D. A. Neisseria meningitidis: an overview of the carrier state. J. Med. Microbiol. 53, 821–832 (2004).
Caugant, D. A., Tzanakaki, G. & Kriz, P. Lessons from meningococcal carriage studies. FEMS Microbiol. Rev. 31, 52–63 (2007).
Jones, C. H. et al. Comparison of phenotypic and genotypic approaches to capsule typing of Neisseria meningitidis by use of invasive and carriage isolate collections. J. Clin. Microbiol. 54, 25–34 (2016).
Tzeng, Y. L., Thomas, J. & Stephens, D. S. Regulation of capsule in Neisseria meningitidis. Crit. Rev. Microbiol. 42, 759–772 (2016).
Ispasanie, E. et al. Spontaneous point mutations in the capsule synthesis locus leading to structural and functional changes of the capsule in serogroup A meningococcal populations. Virulence 9, 1138–1149 (2018).
Claus, H., Maiden, M. C., Maag, R., Frosch, M. & Vogel, U. Many carried meningococci lack the genes required for capsule synthesis and transport. Microbiology 148, 1813–1819 (2002).
Ganesh, K. et al. Molecular characterization of invasive capsule null Neisseria meningitidis in South Africa. BMC Microbiol. 17, 40 (2017).
Neri, A. et al. Carriage meningococcal isolates with capsule null locus dominate among high school students in a non-endemic period, Italy, 2012–2013. Int. J. Med. Microbiol. 309, 182–188 (2019).
Xu, Z., Zhu, B., Xu, L., Gao, Y. & Shao, Z. First case of Neisseria meningitidis capsule null locus infection in China. Infect. Dis. 47, 591–592 (2015).
Diallo, K. et al. Hierarchical genomic analysis of carried and invasive serogroup A Neisseria meningitidis during the 2011 epidemic in Chad. BMC Genom. 18, 398 (2017).
Ren, X. et al. Genomic, transcriptomic, and phenotypic analyses of Neisseria meningitidis isolates from disease patients and their household contacts. mSystems 2, e00127-17 (2017). A comparison of closely related disease- and carriage-associated meningococci using a combination of genetic and phenotypic tools to understand mechanisms associated with virulence.
Sevestre, J. et al. Differential expression of hemoglobin receptor, HmbR, between carriage and invasive isolates of Neisseria meningitidis contributes to virulence: lessons from a clonal outbreak. Virulence 9, 923–929 (2018).
Davidsen, T. & Tønjum, T. Meningococcal genome dynamics. Nat. Rev. Microbiol. 4, 11–22 (2006).
Alfsnes, K. et al. A genomic view of experimental intraspecies and interspecies transformation of a rifampicin-resistance allele into Neisseria meningitidis. Microb. Genom. 4, e000222 (2018).
Ambur, O. H., Frye, S. A. & Tønjum, T. New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J. Bacteriol. 189, 2077–2085 (2007).
Frye, S. A., Nilsen, M., Tønjum, T. & Ambur, O. H. Dialects of the DNA uptake sequence in Neisseriaceae. PLOS Genet. 9, e1003458 (2013).
Lamelas, A. et al. Loss of genomic diversity in a Neisseria meningitidis clone through a colonization bottleneck. Genome Biol. Evol. 10, 2102–2109 (2018).
Maynard Smith, J., Smith, N. H., O'Rourke, M. & Spratt, B. G. How clonal are bacteria? Proc. Natl Acad. Sci. USA 90, 4384–4388 (1993).
Tibayrenc, M. & Ayala, F. J. How clonal are Neisseria species? The epidemic clonality model revisited. Proc. Natl Acad. Sci. USA 112, 8909–8913 (2015).
Achtman, M. & Wagner, M. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 6, 431–440 (2008).
Zhu, P. et al. Fit genotypes and escape variants of subgroup III Neisseria meningitidis during three pandemics of epidemic meningitis. Proc. Natl Acad. Sci. USA 98, 5234–5239 (2001).
Siena, E. et al. In-silico prediction and deep-DNA sequencing validation indicate phase variation in 115 Neisseria meningitidis genes. BMC Genom. 17, 843 (2016).
Saunders, N. J. et al. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol. 37, 207–215 (2000).
Wanford, J. J., Green, L. R., Aidley, J. & Bayliss, C. D. Phasome analysis of pathogenic and commensal Neisseria species expands the known repertoire of phase variable genes, and highlights common adaptive strategies. PLOS ONE 13, e0196675 (2018).
Srikhanta, Y. N., Maguire, T. L., Stacey, K. J., Grimmond, S. M. & Jennings, M. P. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc. Natl Acad. Sci. USA 102, 5547–5551 (2005).
Bayliss, C. D. et al. Neisseria meningitidis escape from the bactericidal activity of a monoclonal antibody is mediated by phase variation of lgtG and enhanced by a mutator phenotype. Infect. Immun. 76, 5038–5048 (2008).
Green, L. R. et al. Phase variation of NadA in invasive Neisseria meningitidis isolates impacts on coverage estimates for 4C-MenB, a MenB vaccine. J. Clin. Microbiol. 56, e00204 (2018).
Tauseef, I. et al. Influence of the combination and phase variation status of the haemoglobin receptors HmbR and HpuAB on meningococcal virulence. Microbiology 157, 1446–1456 (2011).
Lucidarme, J. et al. The distribution and ‘in vivo’ phase variation status of haemoglobin receptors in invasive meningococcal serogroup B disease: genotypic and phenotypic analysis. PLOS ONE 8, e76932 (2013).
Lewis, L. A. et al. Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2. Mol. Microbiol. 32, 977–989 (1999).
Richardson, A. R. & Stojiljkovic, I. Mismatch repair and the regulation of phase variation in Neisseria meningitidis. Mol. Microbiol. 40, 645–655 (2001).
Martin, P., Sun, L., Hood, D. W. & Moxon, E. R. Involvement of genes of genome maintenance in the regulation of phase variation frequencies in Neisseria meningitidis. Microbiology 150, 3001–3012 (2004).
Green, L. R. et al. Potentiation of phase variation in multiple outer membrane proteins during spread of the hyperinvasive Neisseria meningitidis serogroup W ST-11 lineage. J. Infect. Dis. 220, 1109–1117 (2019).
Bickle, T. A. & Krüger, D. H. Biology of DNA restriction. Microbiol. Rev. 57, 434–450 (1993).
Atack, J. M., Yang, Y., Seib, K. L., Zhou, Y. & Jennings, M. P. A survey of type III restriction-modification systems reveals numerous, novel epigenetic regulators controlling phase-variable regulons; phasevarions. Nucleic Acids Res. 46, 3532–3542 (2018).
Seib, K. L., Jen, F. E., Scott, A. L., Tan, A. & Jennings, M. P. Phase variation of DNA methyltransferases and the regulation of virulence and immune evasion in the pathogenic Neisseria. Pathog. Dis. 75, ftx080 (2017). A review describing the importance of epigenetic changes mediated by phase variable DNA methlytransferases in the pathogenic Neisseria species.
Srikhanta, Y. N., Fox, K. L. & Jennings, M. P. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat. Rev. Microbiol. 8, 196–206 (2010).
Phillips, Z. N., Husna, A. U., Jennings, M. P., Seib, K. L. & Atack, J. M. Phasevarions of bacterial pathogens - phase-variable epigenetic regulators evolving from restriction-modification systems. Microbiology 165, 917–928 (2019).
Tan, A. et al. Distribution of the type III DNA methyltransferases modA, modB and modD among Neisseria meningitidis genotypes: implications for gene regulation and virulence. Sci. Rep. 6, 21015 (2016).
Srikhanta, Y. N. et al. Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLOS Pathog. 5, e1000400 (2009).
Jen, F. E., Seib, K. L. & Jennings, M. P. Phasevarions mediate epigenetic regulation of antimicrobial susceptibility in Neisseria meningitidis. Antimicrob. Agents Chemother. 58, 4219–4221 (2014).
Seib, K. L. et al. Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N6-adenine DNA methyltransferases of Neisseria meningitidis. Nucleic Acids Res. 43, 4150–4162 (2015).
Bhat, A. H., Maity, S., Giri, K. & Ambatipudi, K. Protein glycosylation: sweet or bitter for bacterial pathogens? Crit. Rev. Microbiol. 45, 82–102 (2019).
Børud, B. et al. Extended glycan diversity in a bacterial protein glycosylation system linked to allelic polymorphisms and minimal genetic alterations in a glycosyltransferase gene. Mol. Microbiol. 94, 688–699 (2014).
Gault, J. et al. Neisseria meningitidis type IV pili composed of sequence invariable pilins are masked by multisite glycosylation. PLOS Pathog. 11, e1005162 (2015).
Mubaiwa, T. D., et al. The sweet side of the pathogenic Neisseria: the role of glycan interactions in colonisation and disease. Pathog. Dis. 75, ftx063 (2017).
Børud, B. et al. Genetic and molecular analyses reveal an evolutionary trajectory for glycan synthesis in a bacterial protein glycosylation system. Proc. Natl Acad. Sci. USA 108, 9643–9648 (2011).
Børud, B., Bårnes, G. K., Brynildsrud, O. B., Fritzsønn, E. & Caugant, D. A. Genotypic and phenotypic characterization of the O-linked protein glycosylation system reveals high glycan diversity in paired meningococcal carriage isolates. J. Bacteriol. 200, e00794–17 (2018).
Didelot, X., Walker, A. S., Peto, T. E., Crook, D. W. & Wilson, D. J. Within-host evolution of bacterial pathogens. Nat. Rev. Microbiol. 14, 150–162 (2016).
Klughammer, J. et al. Comparative genome sequencing reveals within-host genetic changes in Neisseria meningitidis during invasive disease. PLOS ONE 12, e0169892 (2017). An ultra-deep sequencing analysis of throat-blood strain pairs isolated from four patients detected mutations affecting predominantly contingency genes involved in type IV pilus biogenesis.
Lees, J. A. et al. Large scale genomic analysis shows no evidence for pathogen adaptation between the blood and cerebrospinal fluid niches during bacterial meningitis. Microb. Genom. 3, e000103 (2017).
Omer, H. et al. Genotypic and phenotypic modifications of Neisseria meningitidis after an accidental human passage. PLOS ONE 6, e17145 (2011).
Alamro, M. et al. Phase variation mediates reductions in expression of surface proteins during persistent meningococcal carriage. Infect. Immun. 82, 2472–2484 (2014).
Bårnes, G. K. et al. Whole genome sequencing reveals within-host genetic changes in paired meningococcal carriage isolates from Ethiopia. BMC Genom. 18, 407 (2017).
Bille, E. et al. A chromosomally integrated bacteriophage in invasive meningococci. J. Exp. Med. 201, 1905–1913 (2005).
Bille, E. et al. Association of a bacteriophage with meningococcal disease in young adults. PLOS ONE 3, e3885 (2008).
Siena, E., Bodini, M. & Medini, D. Interplay between virulence and variability factors as a potential driver of invasive meningococcal disease. Comput. Struct. Biotechnol. J. 16, 61–69 (2018). This article supports the hypothesis that invasive meningococcal disease arises from the ability of the bacterium to develop phenotypic variants through stochastic assortment of a repertoire of virulence factors.
Schoen, C., Kischkies, L., Elias, J. & Ampattu, B. J. Metabolism and virulence in Neisseria meningitidis. Front. Cell. Infect. Microbiol. 4, 114 (2014).
Müller, M. G., Ing, J. Y., Cheng, M. K. W., Flitter, B. A. & Moe, G. R. Identification of a phage encoded Ig-binding protein from invasive Neisseria meningitidis. J. Immunol. 191, 3287–3296 (2013).
Brynildsrud, O. B. et al. Acquisition of virulence genes by a carrier strain gave rise to the ongoing epidemics of meningococcal disease in West Africa. Proc. Natl Acad. Sci. USA 115, 5510–5515 (2018). A study showing how acquisition of a few virulence genes by a benign meningococcal carrier strain can have major public health consequences.
Dull, P. M. & McIntosh, E. D. Meningococcal vaccine development – from glycoconjugates against MenACWY to proteins against MenB – potential for broad protection against meningococcal disease. Vaccine 30, B18–B25 (2012).
Vella, M. & Pace, D. Glycoconjugate vaccines: an update. Expert Opin. Biol. Ther. 15, 529–546 (2015).
Chen, W. H. et al. Safety and immunogenicity of a pentavalent meningococcal conjugate vaccine containing serogroups A, C, Y, W, and X in healthy adults: a phase 1, single-centre, double-blind, randomised, controlled study. Lancet Infect. Dis. 18, 1088–1096 (2018).
Christodoulides, M. & Heckels, J. Novel approaches to Neisseria meningitidis vaccine design. Pathog. Dis. 75, ftx033 (2017).
Sette, A. & Rappuoli, R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity 33, 530–541 (2010).
Rappuoli, R., Pizza, M., Masignani, V. & Vadivelu, K. Meningococcal B vaccine (4CMenB): the journey from research to real world experience. Expert Rev. Vaccines 17, 1111–1121 (2018).
Murphy, E. et al. Sequence diversity of the factor H binding protein vaccine candidate in epidemiologically relevant strains of serogroup B Neisseria meningitidis. J. Infect. Dis. 200, 379–389 (2009).
Perez, J. L. et al. From research to licensure and beyond: clinical development of MenB-FHbp, a broadly protective meningococcal B vaccine. Expert Rev. Vaccines 17, 461–477 (2018).
Hoiseth, S. K. et al. A multi-country evaluation of Neisseria meningitidis serogroup B factor H-binding proteins and implications for vaccine coverage in different age groups. Pediatr. Infect. Dis. J. 32, 1096–1101 (2013).
Rodrigues, C. M. C. et al. Genomic surveillance of 4CMenB vaccine antigenic variants among disease-causing Neisseria meningitidis isolates, United Kingdom, 2010–2016. Emerg. Infect. Dis. 24, 673–682 (2018).
Murray, E. G. D. Meningococcus infection of the male urogenital tract and the liability to confusuion with gonococcus infection. Urol. Cutan. Rev. 43, 739–741 (1939).
Givan, K. F., Thomas, B. W. & Johnston, A. G. Isolation of Neisseria meningitidis from the urethra, cervix, and anal canal: further observations. Br. J. Vener. Dis. 53, 109–112 (1977).
Urra, E. et al. Orogenital transmission of Neisseria meningitidis serogroup C confirmed by genotyping techniques. Eur. J. Clin. Microbiol. Infect. Dis. 24, 51–53 (2005).
Ito, S. et al. Male non-gonococcal urethritis: from microbiological etiologies to demographic and clinical features. Int. J. Urol. 23, 325–331 (2016).
Bazan, J. A. et al. Large cluster of Neisseria meningitidis urethritis in Columbus, Ohio, 2015. Clin. Infect. Dis. 65, 92–99 (2017).
Toh, E. et al. Neisseria meningitidis ST11 complex isolates associated with nongonococcal urethritis, Indiana, USA, 2015–2016. Emerg. Infect. Dis. 23, 336–339 (2017).
Ma, K. C. et al. Genomic characterization of urethritis-associated Neisseria meningitidis shows that a wide range of N. meningitidis strains can cause urethritis. J. Clin. Microbiol. 55, 3374–3383 (2017).
Taha, M. K. et al. Evolutionary events associated with an outbreak of meningococcal disease in men who have sex with men. PLOS ONE 11, e0154047 (2016).
Genders, R. E., Spitaels, D., Jansen, C. L., van den Akker, T. W. & Quint, K. D. A misleading urethral smear with polymorphonuclear leucocytes and intracellular diplococci; case report of urethritis caused by Neisseria meningitidis. J. Med. Microbiol. 62, 1905–1906 (2013).
Tzeng, Y. L. et al. Heteroresistance to the model antimicrobial peptide polymyxin B in the emerging Neisseria meningitidis lineage 11.2 urethritis clade: mutations in the pilMNOPQ operon. Mol. Microbiol. 111, 254–268 (2019).
Hill, D. J., Griffiths, N. J., Borodina, E. & Virji, M. Cellular and molecular biology of Neisseria meningitidis colonization and invasive disease. Clin. Sci. 118, 547–564 (2010).
Massari, P. et al. Cutting edge: immune stimulation by neisserial porins is Toll-like receptor 2 and MyD88 dependent. J. Immunol. 168, 1533–1537 (2002).
Woodhams, K. L., Chan, J. M., Lenz, J. D., Hackett, K. T. & Dillard, J. P. Peptidoglycan fragment release from Neisseria meningitidis. Infect. Immun. 81, 3490–3498 (2013).
Bennett, J. S. et al. A genomic approach to bacterial taxonomy: an examination and proposed reclassification of species within the genus Neisseria. Microbiology 158, 1570–1580 (2012).
Katoh, K. & Standley, M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Bryant, D. & Moulton, V. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21, 255–265 (2004).
Huson, D. H. & Bryant, D. User manual for SplitsTree4 V4. 13.1. (2010).
Budroni, S. et al. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc. Natl Acad. Sci. USA 108, 4494–4499 (2011).
Claus, H. et al. Genetic analysis of meningococci carried by children and young adults. J. Infect. Dis. 191, 1263–1271 (2005).
Bentley, S. D. et al. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLOS Genet. 3, e23 (2007).
Parkhill, J. et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404, 502–506 (2000).
Tettelin, H. et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287, 1809–1815 (2000).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Microbiology thanks C. D. Bayliss, D. S. Stephens and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
NeisseriaPubMLST database: https://pubmlst.org/neisseria/
Glossary
- Meninges
-
The membranes surrounding the brain and spinal cord.
- Fulminant
-
Coming on suddenly and with great severity.
- Core genome
-
The set of genes that are present in all (or nearly all) strains of a species or population.
- Dental calculus
-
A form of hardened dental plaque that is caused by precipitation of minerals from saliva and gingival fluid on the teeth.
- Multilocus enzyme electrophoresis
-
A method for characterizing organisms by the relative mobilities under electrophoresis of a large number of intracellular enzymes.
- Multilocus sequence typing
-
A procedure to characterize microbial isolates using the DNA sequences of internal fragments of multiple housekeeping genes.
- DNA uptake sequences
-
Small repeated sequences that are required for DNA binding or uptake in natural transformation in members of the genus Neisseria.
- Pan-genome
-
The sum of genes that are found in at least one strain of a species or population. In addition to the core genome, this includes the accessory genome, which contains dispensable genes present in a subset of the strains.
- Simple sequence repeats
-
DNA tracts in which a short base pair motif is repeated several times, which can be found within the open reading frame or within the promoter region of a gene.
Rights and permissions
About this article
Cite this article
Caugant, D.A., Brynildsrud, O.B. Neisseria meningitidis: using genomics to understand diversity, evolution and pathogenesis. Nat Rev Microbiol 18, 84–96 (2020). https://doi.org/10.1038/s41579-019-0282-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-019-0282-6
This article is cited by
-
Native microbiome dominates over host factors in shaping the probiotic genetic evolution in the gut
npj Biofilms and Microbiomes (2023)
-
Primary septic arthritis of the knee caused by Neisseria meningitidis serogroup B in an elderly patient. Case report and review of the literature
Infection (2023)
-
Click-correlative light and electron microscopy (click-AT-CLEM) for imaging and tracking azido-functionalized sphingolipids in bacteria
Scientific Reports (2021)