- 1Fortiphyte Inc., Berkeley, CA, United States
- 2Innovative Genomics Institute, University of California, Berkeley, Berkeley, CA, United States
- 3Department of Plant Pathology, University of Wisconsin–Madison, Madison, WI, United States
- 4IFAS, Gulf Coast Research and Education Center, University of Florida, Wimauma, FL, United States
- 5Department of Plant Pathology, North Dakota State University, Fargo, ND, United States
Xanthomonas species, Pseudomonas syringae and Ralstonia species are bacterial plant pathogens that cause significant yield loss in many crop species. Generating disease-resistant crop varieties can provide a more sustainable solution to control yield loss compared to chemical methods. Plant immune receptors encoded by nucleotide−binding, leucine−rich repeat (NLR) genes typically confer resistance to pathogens that produce a cognate elicitor, often an effector protein secreted by the pathogen to promote virulence. The diverse sequence and presence/absence variation of pathogen effector proteins within and between pathogen species usually limits the utility of a single NLR gene to protecting a plant from a single pathogen species or particular strains. The NLR protein Recognition of XopQ 1 (Roq1) was recently identified from the plant Nicotiana benthamiana and mediates perception of the effector proteins XopQ and HopQ1 from Xanthomonas and P. syringae respectively. Unlike most recognized effectors, alleles of XopQ/HopQ1 are highly conserved and present in most plant pathogenic strains of Xanthomonas and P. syringae. A homolog of XopQ/HopQ1, named RipB, is present in most Ralstonia strains. We found that Roq1 confers immunity to Xanthomonas, P. syringae, and Ralstonia when expressed in tomato. Strong resistance to Xanthomonas perforans was observed in three seasons of field trials with both natural and artificial inoculation. The Roq1 gene can therefore be used to provide safe, economical, and effective control of these pathogens in tomato and other crop species and reduce or eliminate the need for traditional chemical controls.
Introduction
Bacterial pathogens from the species Pseudomonas syringae and the genera Ralstonia and Xanthomonas can infect many different crop species and inflict significant yield losses when environmental conditions favor disease. Xanthomonas and P. syringae tend to enter plant stem, leaf, or flower tissue through wounds or natural openings, such as stomata or hydathodes, whereas Ralstonia is soilborne, entering roots through wounds and natural openings before colonizing xylem tissue (Vasse et al., 1995; Gudesblat et al., 2009). Once inside the host these bacteria manipulate host metabolism and suppress plant immunity using multiple strategies, including effector proteins delivered by the type III secretion system (Kay and Bonas, 2009; Peeters et al., 2013; Xin et al., 2018). This enables the pathogens to multiply to high titers while the plant tissue is still alive and showing few or no visual symptoms. Once the bacteria reach high populations, they typically cause necrosis of infected leaf tissue or wilting and eventual death of the plant.
Effective control measures for bacterial pathogens are relatively limited, particularly once plants become infected (Davis et al., 2013). Soil fumigation can reduce Ralstonia populations in the soil but this is expensive, potentially hazardous to workers and the environment, and of limited efficacy (Yuliar et al., 2015). Copper sulfate and antibiotics such as streptomycin have been used to control Xanthomonas species and P. syringae but have adverse environmental impacts and many strains have evolved tolerance to these chemicals (Kennelly et al., 2007; Griffin et al., 2017). Applying chemicals that induce systemic acquired resistance, such as acibenzolar-S-methyl, can provide partial control but increases production cost and can depress crop yields when used repeatedly (de Pontes et al., 2016).
The most effective, economical, and safe way to control bacterial pathogens is to plant crop varieties that are immune to the target pathogen (Jones et al., 2014; Vincelli, 2016). Such immunity is often mediated by plant immune receptor genes. Plants have large families of cell surface and intracellular immune receptor proteins that surveil for the presence of invading pathogens (Zipfel, 2014; Jones et al., 2016). Effector proteins delivered by the bacterial type III secretion system are common elicitors of intracellular plant immune receptors encoded by nucleotide−binding domain and leucine−rich repeat containing (NLR) genes (Li et al., 2015; Jones et al., 2016; Kapos et al., 2019). While effector proteins contribute to virulence on a susceptible host, an immune response is activated in the plant if that plant has the cognate receptor to recognize the effector. NLR genes typically confer strong, dominant resistance to pathogens that deliver the cognate recognized effector protein (Tai et al., 1999; Jones and Dangl, 2006; Boller and He, 2009; Deslandes and Rivas, 2012; Li et al., 2015). Disease-resistant plants can be generated by identifying the appropriate plant immune receptor genes and transferring them into the target crop species (Dangl et al., 2013).
We recently identified the Nicotiana benthamiana immune receptor gene named Recognition of XopQ 1 (Roq1), which appears to be restricted to the genus Nicotiana and contributes to resistance against Xanthomonas spp. and P. syringae (Schultink et al., 2017). The Roq1 protein is a Toll/Interleukin−1 Receptor (TIR) NLR immune receptor that mediates recognition of the Xanthomonas effector protein XopQ and the homologous effector HopQ1 from P. syringae. XopQ is present in most species and strains of Xanthomonas (Ryan et al., 2011) and HopQ1 is present in 62% (290 of 467) sequenced putative pathogenic P. syringae strains (Dillon et al., 2019). XopQ/HopQ1 has homology to nucleoside hydrolases and has been shown to enhance virulence on susceptible hosts (Ferrante and Scortichini, 2009; Li et al., 2013), possibly by altering cytokinin levels or interfering with the activity of host 14-3-3 proteins (Giska et al., 2013; Li et al., 2013; Hann et al., 2014; Teper et al., 2014). The conservation of XopQ/HopQ1 and their importance in virulence suggests that Roq1 has widespread potential to confer resistance to these pathogens in diverse crop species. Indeed, transient expression assays demonstrated that Roq1 can recognize XopQ/HopQ1 alleles from Xanthomonas and P. syringae pathogens of tomato, pepper, rice, citrus, cassava, brassica, and bean (Schultink et al., 2017). However, it was not known if Roq1 can confer disease resistance when expressed in a crop plant.
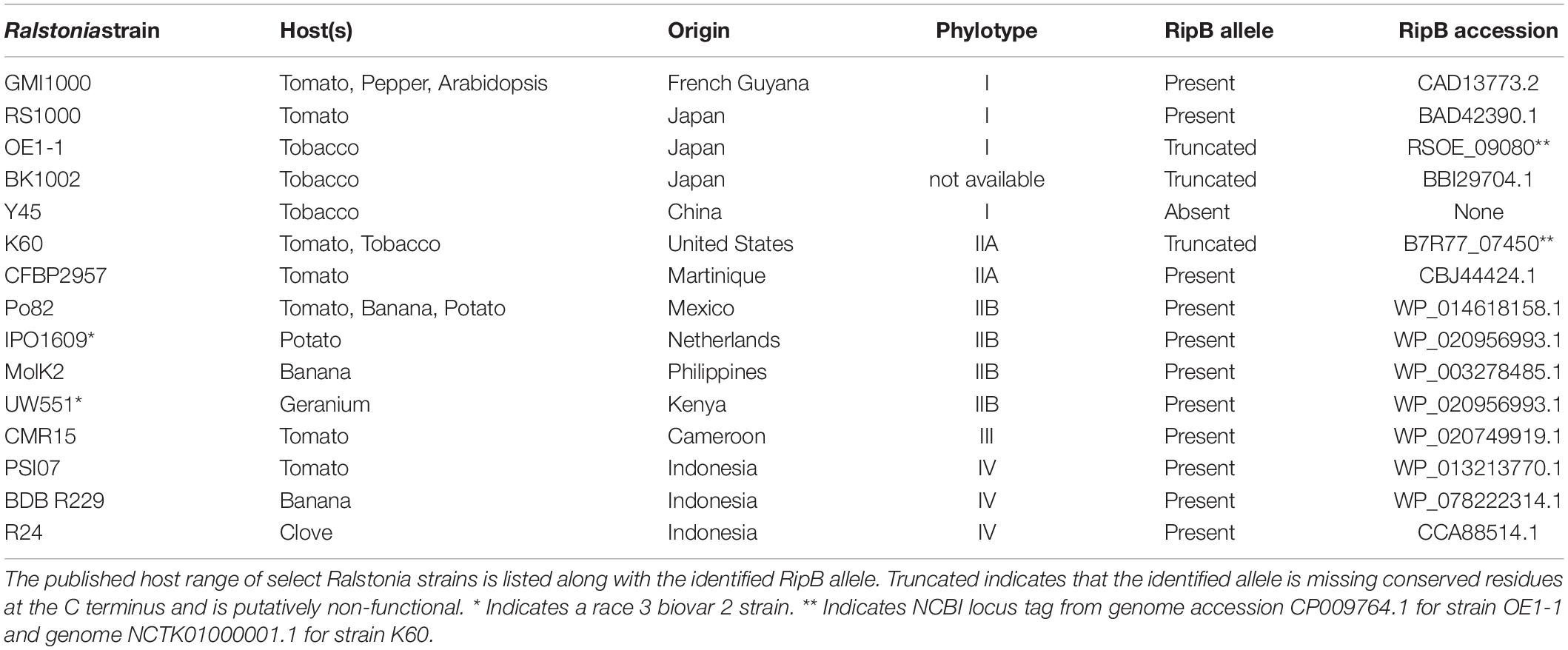
Tomato is one of the most important vegetable crops and is highly susceptible to several bacterial diseases. Bacterial spot, bacterial speck, and bacterial wilt of tomato are caused by Xanthomonas species, P. syringae pv. tomato and Ralstonia, respectively. These diseases are difficult to control, especially if the pathogens become established in a field and environmental conditions favor disease (Rivard et al., 2012; Potnis et al., 2015). Tomato breeding germplasm has only limited resistance against these diseases and in some cases linkage drag has complicated introgression of resistance genes from wild relatives (Sharma and Bhattarai, 2019). Ralstonia contains a homolog of XopQ/HopQ1 called RipB. Roq1 is able to mediate the perception of RipB (Staskawicz and Schultink, 2019), and silencing Roq1 in N. benthamiana resulted in severe wilting phenotypes upon Ralstonia infection (Nakano and Mukaihara, 2019). This suggests that expressing Roq1 in tomato could also confer resistance to bacterial wilt. Like XopQ/HopQ1 in Xanthomonas and P. syringae, RipB is highly conserved and is present in approximately 90% of sequenced Ralstonia isolates (Sabbagh et al., 2019). Here we present data showing that expression of Roq1 in tomato confers resistance against Xanthomonas, Pseudomonas, and Ralstonia upon recognition of the cognate pathogen effector.
Materials and Methods
Generation of Tomato Expressing Roq1
The Roq1 coding sequence was amplified from N. benthamiana cDNA and cloned into the pORE E4 binary plasmid (Coutu et al., 2007). The expression of Roq1 was driven by the constitutive PENTCUP2 promoter, which was derived from tobacco and has been reported to drive expression in leaf, root, and stem tissue (Malik et al., 2002). Agrobacterium tumefaciens co-cultivation was used to transform Roq1 into the tomato variety Fla. 8000 at the University of Nebraska Plant Transformation Core Research Facility. Transformed plants were selected by resistance to kanamycin, confirmed by genotyping, and selfed to obtain homozygous lines.
Bacterial Leaf Spot and Leaf Speck Disease Assays
Xanthomonas cultures were grown in NYG broth (0.5% peptone, 0.3% yeast extract, 2% glycerol) with rifampicin (100 μg/mL) overnight at 30°C. P. syringae cultures were grown in KB broth (1% peptone, 0.15% K2HPO4, 1.5% glycerol, 5 mM MgSO4, pH 7.0) with rifampicin (100 μg/mL) overnight at 28°C. Bacterial cultures were spun down at 5200 g, washed once with 10 mM MgCl2, and then diluted to the appropriate infiltration density with 10 mM MgCl2. Leaf tissue of tomato plants (approximately 4 weeks old) was infiltrated with bacterial solution using a needleless syringe. To quantify bacterial growth, leaf punches were homogenized in water, serially diluted and plated on NYG (for Xanthomonas spp.) or KB (for P. syringae) plates supplemented with 100 μg/mL rifampicin and 50 μg/mL cycloheximide to measure colony forming units. Three biological replicates were performed for each condition and the reported results are representative of at least three independent experiments. Xanthomonas perforans strain 4B, Xanthomonas euvesicatoria strain 85-10, and P. syringae strain DC3000 and the corresponding XopQ/HopQ1 deletion mutants and complemented strains were described previously (Wei et al., 2007; Schwartz et al., 2015; Schultink et al., 2017). The P. syringae pv. tomato Race 1 strain was isolated from a field of tomatoes with the PTO resistance gene in 1993 in California.
Transient Expression of RipB and XopQ
Alleles of RipB from Ralstonia strains GMI1000 and MolK2 (NCBI Genbank accessions CAD13773.2 and WP_003278485) were synthesized and cloned into a BsaI-compatible version of the pORE E4 vector (Coutu et al., 2007). This plasmid was transformed into A. tumefaciens strain C58C1. A. tumefaciens cultures were grown on a shaker overnight at 30°C in LB broth with rifampicin (100 μg/mL), tetracycline (10 μg/mL), and kanamycin (50 μg/mL). The cells were collected by centrifugation and resuspended in infiltration buffer [10 mM 2-(N-morpholino)ethanesulfonic acid, 10 mM MgCl2, pH 5.6], and diluted to an OD600 of 0.5 for infiltration into Nicotiana tabacum leaf tissue. Each experiment was performed on multiple leaves and multiple plants with the selected images being representative of the observed result.
N. tabacum roq1 Mutant Lines
Nicotiana tabacum roq1 mutant lines were generated by transforming N. tabacum with a construct coding for CAS9 and a guide RNA targeting the Roq1 gene with the sequence GATGATAAGGAGTTAAAGAG. This construct was also used for the generation of N. benthamiana roq1 mutants published in Qi et al. (2018). Transformed N. tabacum plants were generated by Agrobacterium co-cultivation and selected for using kanamycin. Transformed plants were genotyped for the presence of mutations at the target site by PCR and Sanger sequencing (Supplementary Table S1).
Bacterial Wilt Virulence Assays
Ralstonia virulence on tomato was measured as previously described (Khokhani et al., 2018). Briefly, cells of Ralstonia strains GMI1000 and UW551 grown overnight in CPG (0.1% casein hydrolysate, 1% peptone, 0.5% glucose, pH 7.0) at 28°C were collected by centrifugation and diluted to an OD600 of 0.1 in water (1 × 108 CFU/mL). 50 mL of this suspension was poured on the soil around 17-day old tomato plants. Disease was rated daily for two weeks on a 0–4 disease index scale, where 0 is no leaves wilted, 1 is 1–25% wilted, 2 is 26–50% wilted, 3 is 51–75% wilted, and 4 is 76–100% wilted. Data represent a total of four biological replicates with 10 plants per replicate. Virulence data were analyzed using repeated measures ANOVA (Khokhani et al., 2018). For petiole infection, the petiole of the first true leaf was cut with a razor blade horizontally approximately 1 cm from the stem. A drop of bacterial solution (2 μL, OD600 = 0.001) was pipetted onto the exposed cut petiole surface.
Field Trial Disease Assays
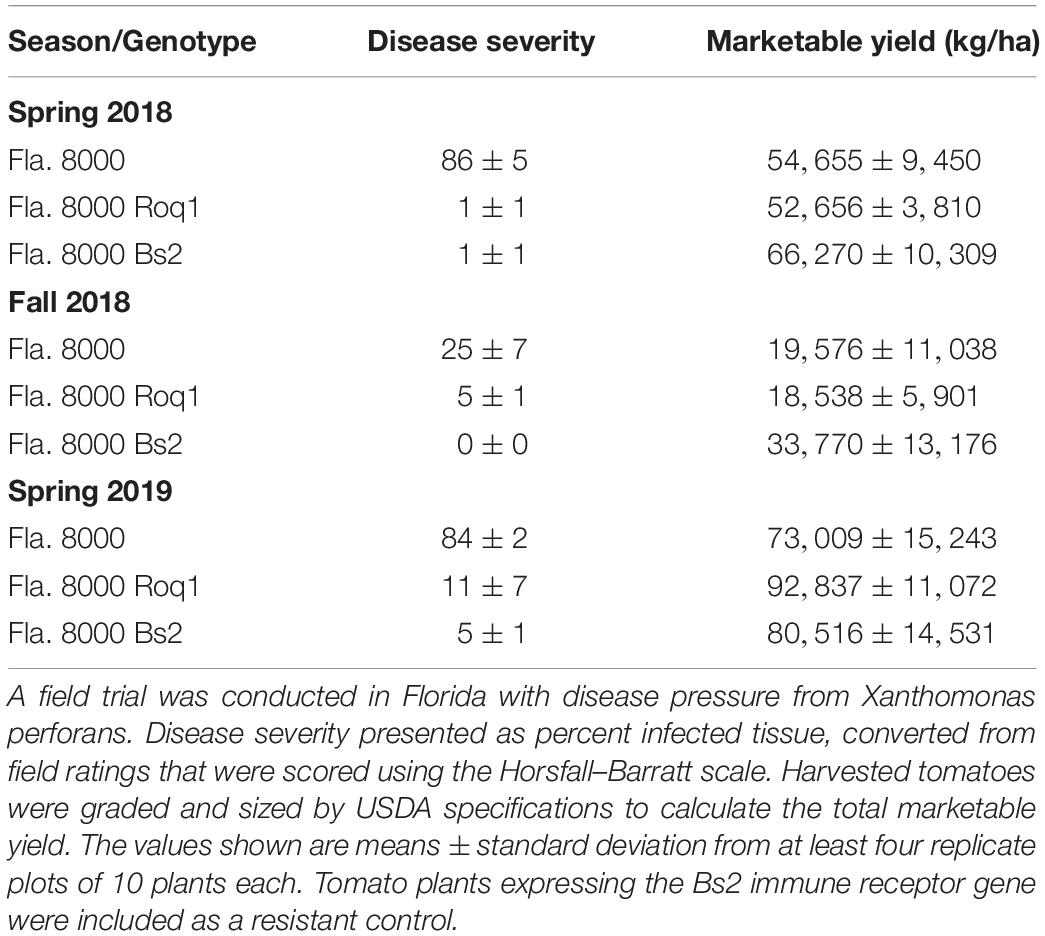
Three field trials were conducted at the University of Florida Gulf Coast Research and Education Center in Balm during the spring seasons of 2018 and 2019 and the fall season of 2018 and under the notification process of the United States Department of Agriculture. Large-fruited, fresh market tomato lines were used in these trials and included the inbred line, Fla. 8000, and nearly isogenic lines containing either Roq1 (event 316.4) or Bs2 (Kunwar et al., 2018). The Roq1 tomato line selected for the field trial was the same line used in the experiments shown in Figures 1, 2, 5. For each trial, seeds were sown directly into peat-lite soilless media (Speedling, Sun City, FL, United States) in 128-cell trays (38 cm3 cell size). Transplants were grown in a greenhouse until 5 or 6 weeks, then planted to field beds that had been fumigated and covered with reflective plastic mulch. Field trials were conducted using a randomized complete block design with four blocks and 10-plant plots. Field plants were staked and tied, and irrigation was applied through drip tape beneath the plastic mulch of each bed. A recommended fertilizer and pesticide program were followed throughout the growing season, excluding the use of plant defense inducers, copper, or other bactericides (Freeman et al., 2018). Fruits were harvested from the inner eight plants of each plot at the breaker stage and beyond and graded for marketability according to USDA specifications with block considered a random effect.
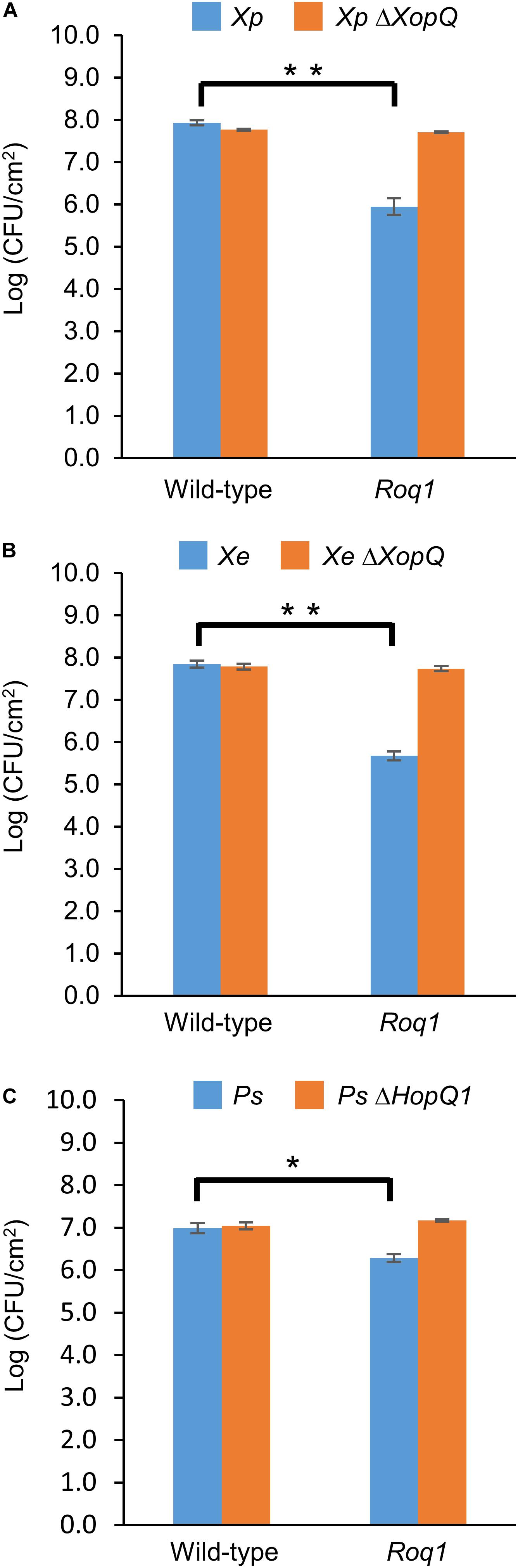
Figure 1. Bacterial growth in tomatoes expressing Roq1. Xanthomonas perforans 4B (Xp), Xanthomonas euvesicatoria 85-10 (Xe), and Pseudomonas syringae DC3000 (Ps) were infiltrated into leaf tissue of wild-type tomato and tomato expressing Roq1 at a low inoculum (OD600 = 0.0001 for Xe and Xp; OD600 = 0.00005 for Ps). Bacterial abundance was quantified by homogenizing leaf punches and counting colony forming units (CFU) per square centimeter of leaf tissue at 6 days post infiltration for Xe and Xp; 3 days post infiltration for Ps. Error bars indicate standard deviation from three biological replicates. *p < 0.05, **p < 0.01 by Student’s t-test.

Figure 2. Bacterial disease symptoms on Roq1 tomato. Xanthomonas perforans 4B (Xp), Xanthomonas euvesicatoria 85-10 (Xe), and Pseudomonas syringae DC3000 (Ps) wild-type and XopQ/HopQ1 knockout strains were infiltrated into tomato leaf tissue at low inoculum and disease symptoms were imaged at 12, 13, and 4 days post infiltration for Xe, Xp, and Ps, respectively. The infiltration was performed using a needleless syringe and circular wounds from the infiltration are visible. The distal part of region of each leaf was infiltrated and the proximal part was left untreated. Xe and Xp were infiltrated at an OD600 of 0.0001 whereas Ps was infiltrated at an OD600 of 0.00005. The images shown here are representative of at least three independent experiments.
Field trials were inoculated with X. perforans race T4 (strain mixture of GEV904, GEV917, GEV1001, and GEV1063). Bacterial strains were grown on nutrient agar medium (BBL, Becton Dickinson and Co., Cockeysville, MD, United States) and incubated at 28°C for 24 h. Bacterial cells were removed from the plates and suspended in a 10 mM MgSO4 solution, and the suspension was adjusted to OD600 = 0.3, which corresponds to 108 CFU/mL. The suspension for each strain was then diluted to 106 CFU/mL, mixed in equal volume, and applied along with polyoxyethylene sorbitan monolaurate (Tween 20; 0.05% [vol/vol]) for field inoculation. Field trial plants were inoculated approximately 3 weeks after transplanting.
Bacterial spot disease severity was recorded three to eight weeks after inoculation using the Horsfall-Barratt scale (Horsfall and Barrat, 1945), and ratings were converted to midpoint percentages for statistical analysis. Blocks were considered random effects.
Generation of the Ralstonia ΔripB Mutant
An unmarked ΔripB mutant was created using sacB selection with the vector pUFR80 (Castañeda et al., 2005). Briefly, the regions upstream and downstream of ripB were amplified using the primers ripBupF/R and ripBdwnF/R (Supplementary Table S1). These fragments were inserted into pUFR80 digested with HindIII and EcoRI using Gibson Assembly (Gibson et al., 2009) (New England Biolabs, Ipswitch, MA, United States) and this construct was incorporated into the genome of strain GMI1000 using natural transformation, with successful integrants selected on CPG + kanamycin (Coupat et al., 2008). Plasmid loss was then selected for on CPG plates containing 5% w/v sucrose. Correct deletions were confirmed using PCR and sequencing.
Phylogenetic Analysis of XopQ, HopQ1, and RipB Alleles
RipB alleles were identified by BLAST search of the NCBI protein database. Clustal Omega (Sievers et al., 2011) was used to generate a multiple sequence alignment with XopQ and HopQ1 alleles. To span the diversity of RipB alleles without having many redundant sequences, only a single sequence was retained if there were multiple identical or nearly identical sequences identified. A maximum likelihood tree was generated using PhyML (Guindon et al., 2010). The phylotype calls of the strains were obtained from previously published worked (Liu et al., 2009; Mukaihara and Tamura, 2009; Safni et al., 2014).
Results
Tomatoes Expressing Roq1 Are Resistant to Xanthomonas and P. syringae
We generated homozygous tomato plants expressing the Roq1 gene from N. benthamiana and tested them for resistance to Xanthomonas and P. syringae by measuring bacterial growth in planta. Population sizes of wild-type X. perforans strain 4B and X. euvesicatoria strain 85-10 were approximately 100-fold smaller in tomato expressing Roq1 compared to wild-type tomato at 6 days post inoculation (Figure 1). In contrast, XopQ deletion mutants multiplied equally well in leaves of both wild-type and Roq1 tomato. Disease symptoms begin as small water-soaked lesions and progress to necrosis of infected tissue. Wild-type X. perforans and X. euvesicatoria caused severe disease symptoms on wild-type tomato plants but failed to cause visible symptoms on Roq1 plants (Figure 2). The XopQ mutants caused similar disease symptoms on both wild-type and Roq1 tomato. Similar results were observed for P. syringae DC3000, and its HopQ1 mutant (Figures 1, 2) and a Race 1 isolate of P. syringae pv. tomato (Supplementary Figure S1). Tomatoes expressing Roq1 were resistant to Xanthomonas and Pseudomonas XopQ/HopQ1 mutants complemented with a wild-type copy of XopQ/HopQ1 (Supplementary Figure S2). A second tomato line expressing Roq1, derived from an independent transformation event, also showed resistance to wild-type X. euvesicatoria and X. perforans but not to the XopQ deletion strains (Supplementary Figure S3).
Expression of Roq1 Confers Resistance to Xanthomonas perforans in the Field
To determine if the resistance observed in growth chamber experiments would hold up under commercial tomato production conditions, we tested the ability of Roq1 tomatoes to resist X. perforans infection in the field. Roq1 tomatoes were grown along with the Fla. 8000 wild-type parent as well as a Fla. 8000 variety expressing the Bs2 gene from pepper as a resistant control (Kunwar et al., 2018). For each of the three growing seasons, both Roq1 and the resistant Bs2 control tomatoes showed significantly lower disease severity than the parental Fla. 8000 variety (Table 1) (p < 0.05). The total marketable yield of the Roq1 plants was not significantly different from that of the susceptible parent for any of the three seasons (p > 0.05). No obvious difference in growth morphology was observed between Roq1 and wild-type tomato plants (Supplementary Figure S4).
The Ralstonia Effector RipB, a Homolog of XopQ/HopQ1, Is Recognized by Roq1
RipB, considered a “core” Ralstonia effector, is present in approximately 90% of sequenced strains (Sabbagh et al., 2019) making it an attractive target ligand for engineering crop plants to be resistant to this pathogen. Roq1 perceives diverse alleles of XopQ and HopQ1 and we hypothesized that it can also recognize different alleles of RipB. We constructed a phylogenetic tree using a subset of RipB alleles identified by BLAST search to approximately span the diversity of this effector in Ralstonia (Figure 3). The Ralstonia genus contains many diverse strains that have been divided into four phylotypes based on based on sequence analysis of the internal transcribed spacer region of the 16S–23S rRNA gene (Poussier et al., 2000; Prior and Fegan, 2004; Safni et al., 2014). We selected RipB alleles from Ralstonia strains GMI1000 and MolK2, from phylotypes I and II, respectively, for subsequent analysis. These two RipB alleles share 71% amino acid identity with each other and approximately 52% identity with XopQ excluding the divergent N terminus containing the putative type III secretion signal. An alignment of these two RipB proteins with XopQ and HopQ1 is shown in Supplementary Figure S5. To test for Roq1-dependent recognition of RipB, we used Agrobacterium to transiently express RipB from GMI1000 and Molk2 in leaf tissue of wild-type and roq1 mutant N. tabacum. The N. tabacum roq1-1 mutant was generated using a CRISPR/CAS9 construct targeting exon 1 of the Roq1 gene (Supplementary Figure S6). Both RipB alleles triggered a strong hypersensitive/cell death response in wild-type N. tabacum, indicating immune activation. This response was absent in the roq1-1 mutant but could be restored by transiently expressing Roq1 along with XopQ, RipBGMI1000, or RipBMolk2 (Figure 4).
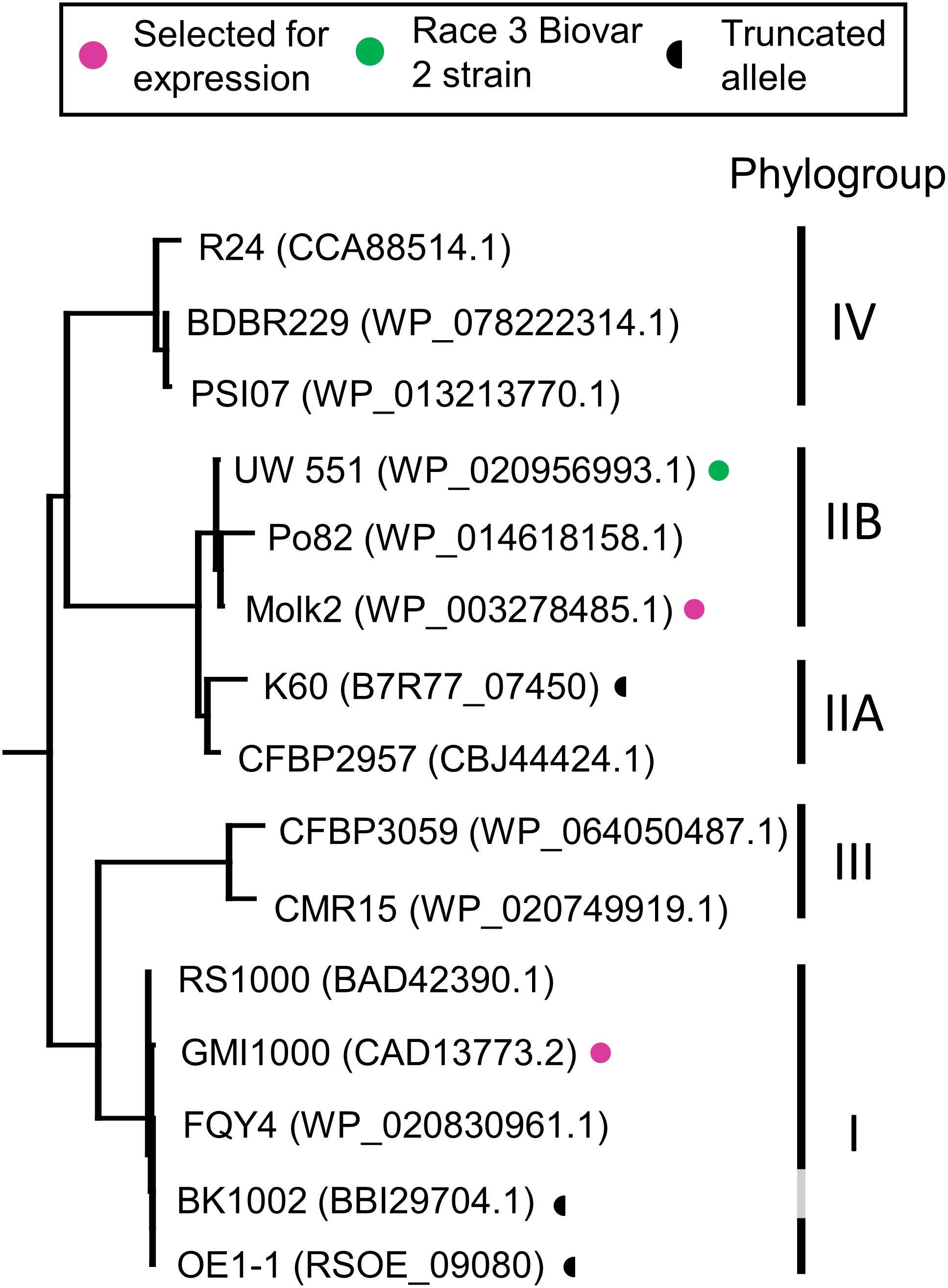
Figure 3. Phylogenetic tree of RipB proteins. RipB alleles from Ralstonia strains GMI1000 and MolK2 were cloned for testing in this study and are indicated by magenta dots. Putatively non-functional RipB alleles with C-terminal truncations are marked with a half circle. Strain names are given along with the associated NCBI protein accession or locus tag. Phylotype groups were labeled based on previous publications with the exception of BK1002, for which an assignment was not available. The tree was rooted using XopQ and HopQ1.
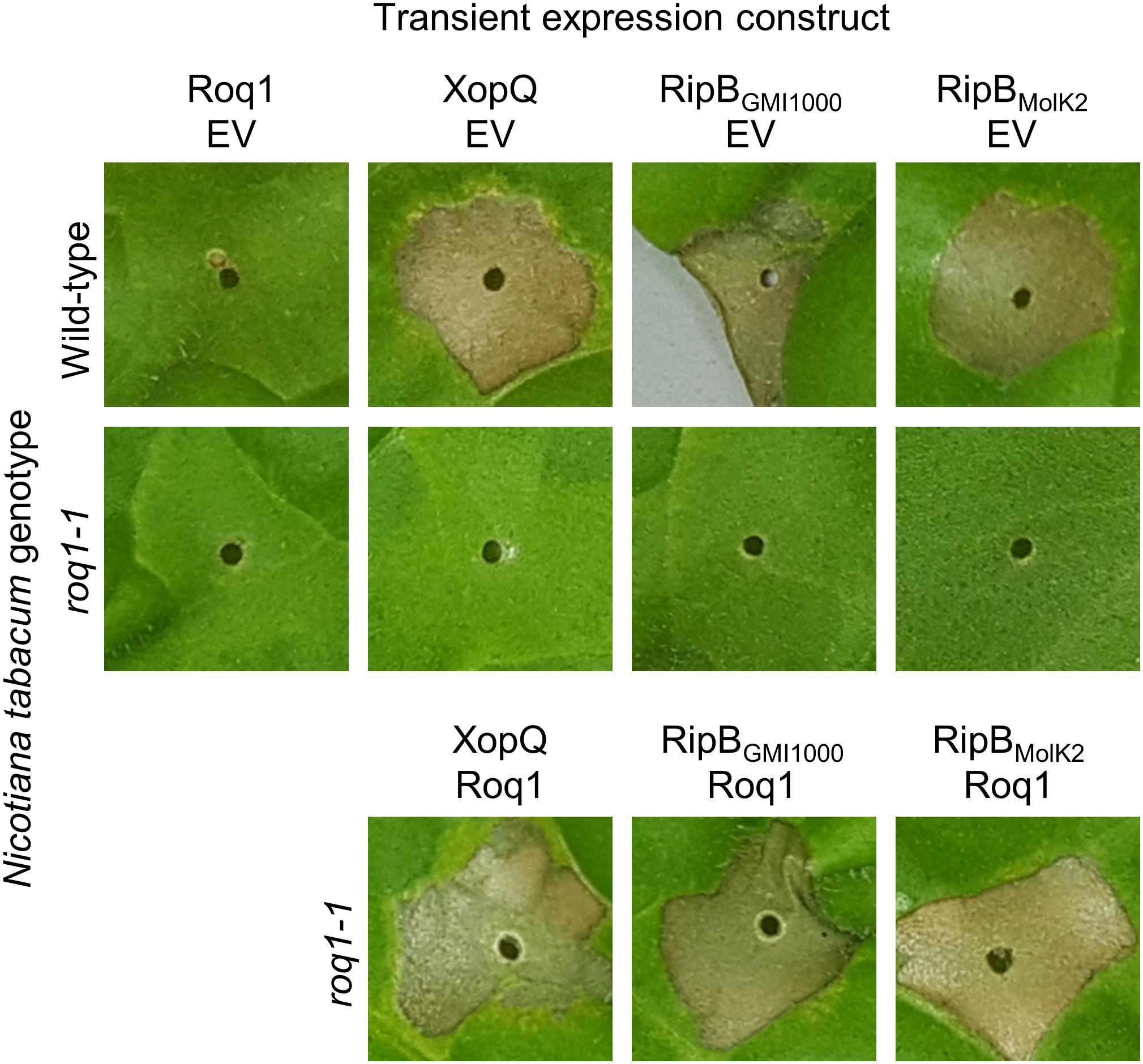
Figure 4. Roq1-dependent recognition of RipB in Nicotiana tabacum. Agrobacterium tumefaciens was used to transiently express XopQ, RipBGMI1000 and RipBMolk2 along with either Roq1 or an empty vector (EV) control in wild-type Nicotiana tabacum and a roq1 loss of function mutant. The Agrobacterium was infiltrated at a total OD600 of 0.5 and the leaves were imaged at 3 days post infiltration. The images shown here are representative of three independent experiments.
Roq1 Tomatoes Are Resistant to Ralstonia Containing RipB
Our observation that Roq1 can recognize RipB in leaf transient expression assays suggested that Roq1 can mediate resistance to bacterial wilt caused by Ralstonia in tomato. We tested this hypothesis by challenging wild-type and Roq1-expressing tomato plants with Ralstonia strain GMI1000 using a soil soak inoculation disease assay. Wild-type plants developed severe wilting approximately 7 days after inoculation, whereas Roq1 tomato plants remained mostly healthy over the 2-week time course (Figure 5A and Supplementary Figure S7). The Roq1 tomato plants were susceptible to a deletion mutant lacking RipB (GMI1000 ΔripB). We also challenged plants by introducing bacteria directly to the xylem by placing bacteria on the surface of a cut petiole. Wild-type plants were wilted by eight days whereas Roq1 plants remained healthy (Figure 5B). Tomatoes expressing Roq1 were also resistant to Ralstonia strain UW551, which is a race 3 biovar 2 potato brown rot strain from phylotype II that has a RipB allele (Figure 3 and Supplementary Figure S8).
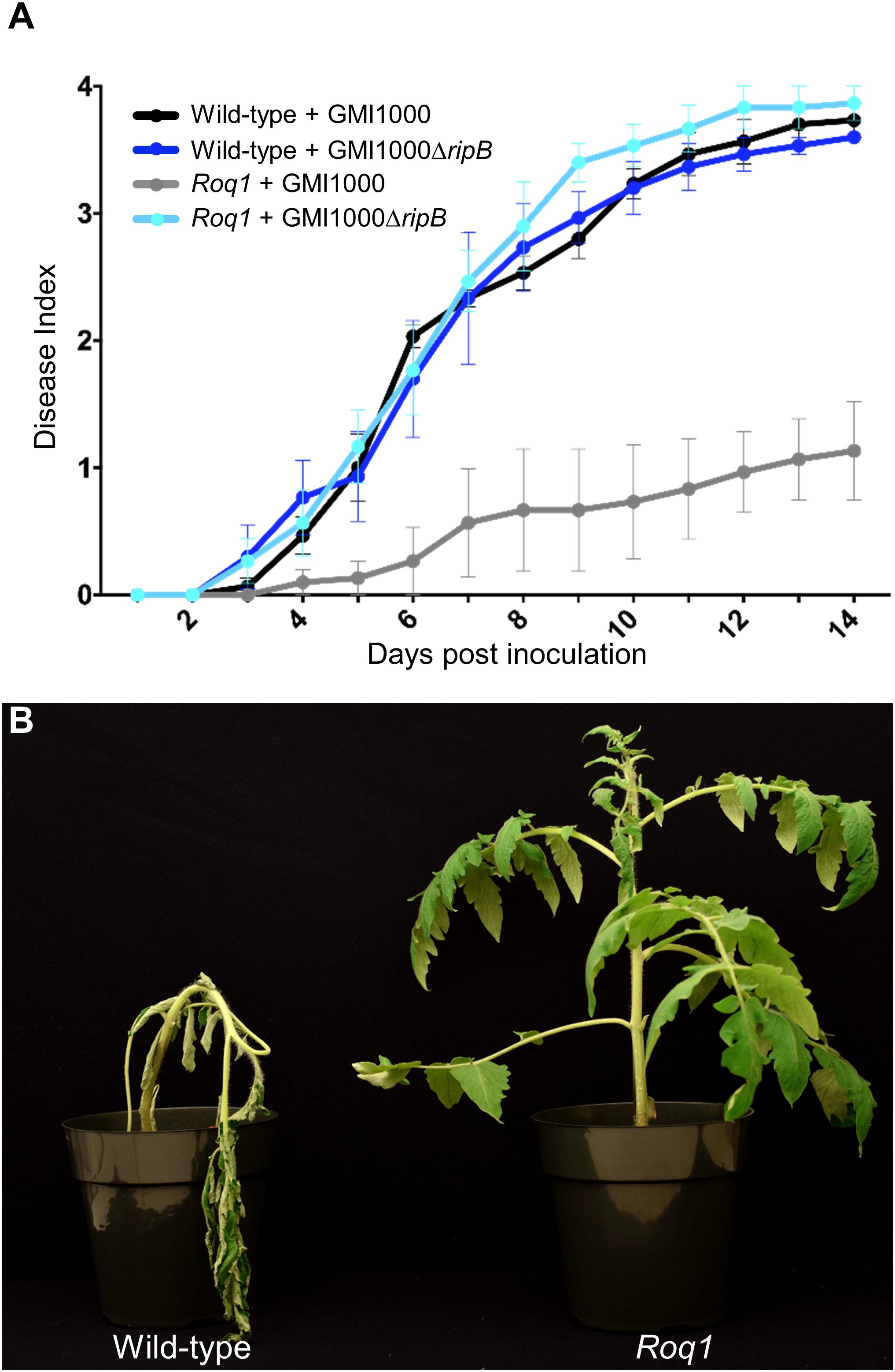
Figure 5. Bacterial Wilt disease development in Roq1 tomatoes. (A) Wild-type and Roq1 tomato plants were infected with wild-type and RipB mutant Ralstonia strain GMI1000 by soil soak inoculation. Disease symptoms were monitored over 14 days, with no wilting corresponding to a Disease Index of 0 and complete wilting corresponding to a Disease Index of 4. Error bars indicate standard error from four biological replicates. Disease progression of wild-type Ralstonia on tomatoes expressing Roq1 was significantly lower (p < 0.05) than the three other conditions (Friedman test with Dunn’s multiple comparisons test). (B) Wild-type and Roq1 tomato plants 8 days after petiole inoculation with approximately 2000 cells of wild-type GMI1000.
Distribution of RipB Alleles in Ralstonia
To investigate the potential for using Roq1 to protect plants from Ralstonia, we investigated the occurrence of RipB alleles in select strains. Table 2 lists some Ralstonia strains with their known hosts along with their respective phylotype and RipB allele accession information. Table 2 illustrates that strains lacking putative functional RipB alleles correlate with strains that are virulent on tobacco, which contains a native Roq1 gene. All strains in Table 2 except for tobacco pathogenic strains K60, Y45, BK1002, and OE1-1 contain putative full-length and functional RipB alleles. Relative to other RipB alleles, the K60 RipB allele is truncated after residue 437 and missing approximately 65 C-terminal residues and the OE1-1 allele is truncated after residue 417, missing approximately 77 residues based on a published genome sequence (Hayes et al., 2017) (Supplementary Figure S5). Y45 does not have a predicted RipB allele based on a draft genome sequence (Li et al., 2011). Published gene models for RipB disagree on which start codon is the correct one, leading some RipB alleles to look like they are missing part of the N terminus or have N-terminal extensions. Analysis of the DNA sequence of diverse RipB alleles showed that out of three possible in-frame start codons, only a single putative start codon is conserved among Ralstonia strains from all four phylotypes (Supplementary Figure S9), suggesting that this is the true start codon and eliminating the N-terminal discrepancy between different RipB alleles.
Discussion
Roq1 expression in tomato confers strong resistance to X. perforans, X. euvesicatoria, and P. syringae pv. tomato. Its effectiveness is dependent on the presence of the recognized effector protein XopQ/HopQ1 (Figures 1, 2). Field trials revealed that tomatoes expressing Roq1 were less susceptible to X. perforans than wild-type tomatoes in conditions approximating commercial production (Table 1). Roq1 conferred a similar level of resistance as the Bs2-containing resistant check variety in one season and was slightly weaker in the other two. Bacterial spot caused by X. perforans can cause lesions on fruits, making them unsuitable for commercial sale, and also reduce plant productivity by damaging leaf tissue. The onset of fruit lesions requires high disease pressure during a particular phase of fruit development. Environmental conditions did not favor the development of fruit lesions and we did not observe significant fruit lesion formation on any of the genotypes in any of the three seasons. Despite showing a strong reduction in foliar disease symptoms, the Roq1 line did not have a significantly greater yield than the susceptible parental variety. A possible explanation for this finding is that bacterial spot did not appear to be a major constraint on yield in any of the three seasons. In spring 2018, weather conditions promoted the development of disease only late in the season after much of the yield was already set. Fall 2018 was unseasonably hot and dry for most of the season resulting in low disease pressure and very poor yield for all genotypes. Of the three seasons, spring 2019 had weather conditions expected to be most conducive for observing an impact of bacterial spot on marketable yield with mid-season rain promoting the development of disease symptoms. The average marketable yield for the Roq1 tomatoes was 27% higher than wild-type in this season, although a relatively small sample size (four replicate plots of 10 plants each) and a large variability of yield between plots resulted in a p-value of 0.08 by Student’s t-test. Notably the yield of the resistant check variety expressing Bs2 was not significantly higher than the susceptible control in this season, though it was previously reported to give a yield increase of 1.5–11x relative to susceptible varieties under high disease pressure (Horvath et al., 2012). This suggests that bacterial spot was not severe enough to have a strong impact on yield in this season and that Roq1 may result in an increase in marketable yield under stronger disease pressure.
It was unclear if Roq1 could confer resistance to Ralstonia because it colonizes different tissues than Xanthomonas and P. syringae. While Xanthomonas and P. syringae colonize tomato leaf tissue, Ralstonia enters through the roots and colonizes xylem vessels. Although the type III secretion system is essential for virulence in Ralstonia, it is not clear when and where the pathogen delivers effectors into host cells. It was therefore not clear if Roq1 would be able to confer resistance to this pathogen in tomato. Here we demonstrated that tomato plants expressing Roq1 had strong resistance to Ralstonia expressing RipB as measured by both soil soak and cut-petiole inoculation assays (Figure 5). In addition to conferring resistance to the phylotype I strain GMI1000, Roq1 also confers resistance to Ralstonia race 3 biovar 3 strain UW551, a phylotype II strain that can overcome other known sources of bacterial wilt resistance in tomato (Milling et al., 2011). Some but not all of the Roq1 tomatoes inoculated with GMI1000 by soil soak were colonized by a moderate or low population of Ralstonia (Supplementary Figure S10). This observation suggests that Roq1-mediated immune responses may act to both restrict the establishment of vascular colonization and separately reduce bacterial titers if colonization does occur. Activation of immune receptors, including Roq1, is known to induce many defense-associated genes with different putative activities (Sohn et al., 2014; Qi et al., 2018), presumably acting to inhibit pathogen virulence by distinct mechanisms. The observation that Roq1 inhibits both colonization establishment and population growth suggests that at least two independent downstream defense responses mediate the observed resistance phenotype.
The Roq1 tomatoes were susceptible to a Ralstonia mutant lacking RipB, indicating that the resistance depends on the interaction between RipB and Roq1. This is consistent with the observation that several naturally occurring Ralstonia strains that can infect tobacco have a truncated or are missing the RipB effector (Table 2) (Nakano and Mukaihara, 2019), suggesting that losing RipB can allow the pathogen to overcome the native Roq1 gene present in N. tabacum. Tobacco-infecting strains K60 and OE1-1 contain independently truncated RipB alleles (Figure 3 and Supplementary Figure S5) and there have likely been multiple independent gene loss events which enable strains to evade Roq1-mediated resistance. Similarly, HopQ1 has been lost in strains of P. syringae that can infect tobacco (Denny, 2006; Ferrante and Scortichini, 2009; Li et al., 2011). This suggests that this effector is not essential for virulence in all circumstances and it would therefore be prudent to deploy Roq1 in combination with other disease resistance traits to avoid resistance breakdown due to pathogens losing XopQ/HopQ1/RipB. Although minor foliar symptoms were observed on the Roq1 tomatoes, particularly in spring 2019 (Table 1), we do not believe this was due to a naturally occurring XopQ mutant arising during the trial. Instead, we think that these low disease scores may have been the result of fungal diseases, which can cause foliar symptoms that look similar to bacterial spot, or by the Roq1 tomatoes supporting a low level of bacterial growth.
No other known NLR immune receptor confers resistance against such a broad range of bacterial pathogenic genera as Roq1. Effectors that are recognized by NLR proteins act as avirulence factors and are under strong evolutionary pressure to diversify or be lost to evade immune activation. Therefore the effector repertoires of pathogens are often quite diverse, with relatively few “core” effectors conserved within a species and even fewer shared between different genera (Grant et al., 2006). Effectors recognized by plant NLRs are typically narrowly conserved within a single bacterial genus (Kapos et al., 2019). One such effector is AvrBs2, recognized by the Bs2 receptor from pepper, which is present in many Xanthomonas strains but is absent from P. syringae and Ralstonia. In contrast, XopQ/HopQ1/RipB is highly conserved in most Xanthomonas, P. syringae, and Ralstonia strains that cause disease in crop plants including kiwi (P. syringae pv. actinidae), banana (Ralstonia and X. campestris pv. musacearum), stone fruit (P. syringae), pepper (X. euvesicatoria), citrus (X. citri), strawberry (X. fragariae), brassica (X. campestris), rice (X. oryzae), potato (Ralstonia), and others. Ralstonia race 3 biovar 2 strains are of particular concern because they are cold tolerant and potentially threaten potato cultivation in temperate climates. As a result, Ralstonia race 3 biovar 2 strains are strictly regulated quarantine pathogens in Europe and North America and is on the United States Select Agent list. The ability of Roq1 to protect tomato from the race 3 biovar 2 strain UW551 (Supplementary Figure S8) suggests that Roq1 can also protect potato from this high-concern pathogen. This work demonstrates the widespread potential of using naturally occurring plant immune receptors to safely, sustainably, and economically manage diverse and difficult to control pathogen species.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
NT and AS wrote the manuscript and performed Pseudomonas and Ralstonia petiole infection assays. AS carried out Xanthomonas infection and Agrobacterium transient expression experiments. UG and SH performed Xanthomonas field experiments. CH constructed the Ralstonia knockout and performed Ralstonia soil soak assays, supervised by CA. All authors analyzed the results and edited and approved the manuscript.
Funding
This work was supported in part by the National Institute of Food and Agriculture, US Department of Agriculture under award number 2016-67012-25106, and the UC Berkeley Innovative Genomics Institute. CH was supported by an NSF Predoctoral Fellowship.
Conflict of Interest
AS and NT are employees of and have a financial stake in Fortiphyte Inc., which has intellectual property rights related to the Roq1 resistance gene.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript has been released as a pre-print at bioRxiv (Thomas et al., 2019). We thank Shirley Sato and Tom Clemente of the University of Nebraska Plant Transformation Core Research Facility for the transformation of tomato. We thank Myeong-Je Cho and Julie Pham of the UC Berkeley Innovative Genomics Institute for transformation of Nicotiana tabacum.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00463/full#supplementary-material
References
Boller, T., and He, S. Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324, 742–744. doi: 10.1126/science.1171647
Castañeda, A., Reddy, J. D., El-Yacoubi, B., and Gabriel, D. W. (2005). Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity, but one avr gene affected race specificity. Mol. Plant. Microbe Interact. 18, 1306–1317.
Coupat, B., Chaumeille-Dole, F., Fall, S., Prior, P., Simonet, P., Nesme, X., et al. (2008). Natural transformation in the Ralstonia solanacearum species complex: number and size of DNA that can be transferred. FEMS Microbiol. Ecol. 66, 14–24. doi: 10.1111/j.1574-6941.2008.00552.x
Coutu, C., Brandle, J., Brown, D., Brown, K., Miki, B., Simmonds, J., et al. (2007). pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res. 16, 771–781. doi: 10.1007/s11248-007-9066-2
Dangl, J. L., Horvath, D. M., and Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. doi: 10.1126/science.1236011
Davis, R. M., Miyao, G., Subbarao, K. V., Stapleton, J. J., and Aegerter, A. B. J. (2013). UC IPM: UC Management Guidelines - TOMATO:Diseases. Available at: http://ipm.ucanr.edu/PMG/r783101611.html (Accessed June 17, 2019).
de Pontes, N. C., Nascimento, A. D. R., Golynski, A., Maffia, L. A., and Rogério de Oliveira, J. (2016). Intervals and number of applications of acibenzolar-S-methyl for the control of bacterial spot on processing tomato. Plant Dis. 100, 2126–2133. doi: 10.1094/pdis-11-15-1286-re
Denny, T. (2006). “Plant pathogenic Ralstonia species,” in Plant-Associated Bacteria, ed. S. S. Gnanamanickam (Dordrecht: Springer), 573–644. doi: 10.1007/1-4020-4538-7_16
Deslandes, L., and Rivas, S. (2012). Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 17, 644–655. doi: 10.1016/j.tplants.2012.06.011
Dillon, M. M., Almeida, R. N. D., Laflamme, B., Martel, A., Weir, B. S., Desveaux, D., et al. (2019). Molecular evolution of Pseudomonas syringae Type III secreted effector proteins. Front. Plant Sci. 10:418.
Ferrante, P., and Scortichini, M. (2009). Identification of Pseudomonas syringae pv. actinidiae as causal agent of bacterial canker of yellow kiwifruit (Actinidia chinensis Planchon) in Central Italy. J. Phytopathol. 157, 768–770. doi: 10.1111/j.1439-0434.2009.01550.x
Freeman, J. H., McAvoy, E. J., Boyd, N. S., Kanissery, R., Smith, H. A., Desaeger, J., et al. (2018). “Tomato production,” in Vegetable Production Handbook of Florida 2018–2019, eds H. A. Smith, J. H. Freeman, P. J. Dittmar, M. L. Paret, and G. E. Vallad (Lincolnshire, IL: Vance Publishing Corporation), 349–393.
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A. III, and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Giska, F., Lichocka, M., Piechocki, M., Dadlez, M., Schmelzer, E., Hennig, J., et al. (2013). Phosphorylation of HopQ1, a type III effector from Pseudomonas syringae, creates a binding site for host 14-3-3 proteins. Plant Physiol. 161, 2049–2061. doi: 10.1104/pp.112.209023
Grant, S. R., Fisher, E. J., Chang, J. H., Mole, B. M., and Dangl, J. L. (2006). Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60, 425–449. doi: 10.1146/annurev.micro.60.080805.142251
Griffin, K., Gambley, C., Brown, P., and Li, Y. (2017). Copper-tolerance in Pseudomonas syringae pv. tomato and Xanthomonas spp. and the control of diseases associated with these pathogens in tomato and pepper. A systematic literature review. Crop Protoc. 96, 144–150. doi: 10.1016/j.cropro.2017.02.008
Gudesblat, G. E., Torres, P. S., and Vojnov, A. A. (2009). Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell-to-cell signal-regulated virulence factor. Plant Physiol. 149, 1017–1027. doi: 10.1104/pp.108.126870
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hann, D. R., Domínguez-Ferreras, A., Motyka, V., Dobrev, P. I., Schornack, S., Jehle, A., et al. (2014). The Pseudomonas type III effector HopQ1 activates cytokinin signaling and interferes with plant innate immunity. New Phytol. 201, 585–598. doi: 10.1111/nph.12544
Hayes, M. M., MacIntyre, A. M., and Allen, C. (2017). Complete genome sequences of the plant pathogens Ralstonia solanacearum Type Strain K60 and R. solanacearum race 3 biovar 2 strain UW551. Genome Announc. 5:e01088-17. doi: 10.1128/genomeA.01088-17
Horsfall, J. G., and Barrat, R. W. (1945). An improved grading system for measuring plant diseases. Phytopathology 35:655.
Horvath, D. M., Stall, R. E., Jones, J. B., Pauly, M. H., Vallad, G. E., Dahlbeck, D., et al. (2012). Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PloS One 7:e42036. doi: 10.1371/journal.pone.0042036
Jones, J. D. G., Vance, R. E., and Dangl, J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354, doi: 10.1126/science.aaf6395
Jones, J. D. G., Witek, K., Verweij, W., Jupe, F., Cooke, D., Dorling, S., et al. (2014). Elevating crop disease resistance with cloned genes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130087. doi: 10.1098/rstb.2013.0087
Kapos, P., Devendrakumar, K. T., and Li, X. (2019). Plant NLRs: from discovery to application. Plant Sci. 279, 3–18. doi: 10.1016/j.plantsci.2018.03.010
Kay, S., and Bonas, U. (2009). How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 12, 37–43. doi: 10.1016/j.mib.2008.12.006
Kennelly, M. M., Cazorla, F. M., de Vicente, A., Ramos, C., and Sundin, G. W. (2007). Pseudomonas syringae diseases of fruit trees: progress toward understanding and control. Plant Dis. 91, 4–17. doi: 10.1094/pd-91-0004
Khokhani, D., Tran, T. M., Lowe-Power, T. M., and Allen, C. (2018). Plant assays for quantifying Ralstonia solanacearum virulence. Bio Protocol 8:3028.
Kunwar, S., Iriarte, F., Fan, Q., Evaristo da Silva, E., Ritchie, L., Nguyen, N. S., et al. (2018). Transgenic expression of EFR and Bs2 genes for field management of bacterial wilt and bacterial spot of tomato. Phytopathology 108, 1402–1411. doi: 10.1094/phyto-12-17-0424-r
Li, W., Yadeta, K. A., Elmore, J. M., and Coaker, G. (2013). The Pseudomonas syringae effector HopQ1 promotes bacterial virulence and interacts with tomato 14-3-3 proteins in a phosphorylation-dependent manner. Plant Physiol. 161, 2062–2074. doi: 10.1104/pp.112.211748
Li, X., Kapos, P., and Zhang, Y. (2015). NLRs in plants. Curr. Opin. Immunol. 32, 114–121. doi: 10.1016/j.coi.2015.01.014
Li, Z., Wu, S., Bai, X., Liu, Y., Lu, J., Liu, Y., et al. (2011). Genome sequence of the tobacco bacterial wilt pathogen Ralstonia solanacearum. J. Bacteriol. 193, 6088–6089. doi: 10.1128/jb.06009-11
Liu, Y., Kanda, A., Kiba, A., Hikichi, Y., and Ohnishi, K. (2009). Distribution of avirulence genes avrA and popP1 in 22 Japanese phylotype I strains of Ralstonia solanacearum. J. Gen. Plant Pathol. 75, 362–368. doi: 10.1007/s10327-009-0189-6
Malik, K., Wu, K., Li, X. Q., Martin-Heller, T., Hu, M., Foster, E., et al. (2002). A constitutive gene expression system derived from the tCUP cryptic promoter elements. Theor. Appl. Genet. 105, 505–514. doi: 10.1007/s00122-002-0926-0
Milling, A., Babujee, L., and Allen, C. (2011). Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants. PLoS One 6:e15853. doi: 10.1371/journal.pone.0015853
Mukaihara, T., and Tamura, N. (2009). Identification of novel Ralstonia solanacearum type III effector proteins through translocation analysis of hrpB-regulated gene products. Microbiology 155, 2235–2244. doi: 10.1099/mic.0.027763-0
Nakano, M., and Mukaihara, T. (2019). The type III effector RipB from Ralstonia solanacearum RS1000 acts as a major avirulence factor in Nicotiana benthamiana and other Nicotiana species. Mol. Plant Pathol. 20, 1237–1251. doi: 10.1111/mpp.12824
Peeters, N., Carrère, S., Anisimova, M., Plener, L., Cazalé, A.-C., and Genin, S. (2013). Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics 14:859. doi: 10.1186/1471-2164-14-859
Potnis, N., Timilsina, S., Strayer, A., Shantharaj, D., Barak, J. D., Paret, M. L., et al. (2015). Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol. Plant Pathol. 16, 907–920. doi: 10.1111/mpp.12244
Poussier, S., Prior, P., Luisetti, J., Hayward, C., and Fegan, M. (2000). Partial sequencing of the hrpB and endoglucanase genes confirms and expands the known diversity within the Ralstonia solanacearum species complex. Syst. Appl. Microbiol. 23, 479–486. doi: 10.1016/s0723-2020(00)80021-1
Prior, P., and Fegan, M. (2004). “Recent developments in the phylogeny and classification of Ralstonia solanacearum,” in Proceedings of the I International Symposium on Tomato Diseases 695 (Leuven: International Society for Horticultural Science), 127–136. doi: 10.17660/actahortic.2005.695.14
Qi, T., Seong, K., Thomazella, D. P. T., Kim, J. R., Pham, J., Seo, E., et al. (2018). NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc. Natl. Acad. Sci. U.S.A. 115, E10979–E10987.
Rivard, C. L., O’Connell, S., Peet, M. M., Welker, R. M., and Louws, F. J. (2012). Grafting tomato to manage bacterial wilt caused by Ralstonia solanacearum in the Southeastern united states. Plant Dis. 96, 973–978. doi: 10.1094/pdis-12-10-0877
Ryan, R. P., Vorhölter, F.-J., Potnis, N., Jones, J. B., Van Sluys, M.-A., Bogdanove, A. J., et al. (2011). Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat. Rev. Microbiol. 9, 344–355. doi: 10.1038/nrmicro2558
Sabbagh, C. R. R., Carrère, S., Lonjon, F., Vailleau, F., Macho, A. P., Genin, S., et al. (2019). Pangenomic type III effector database of the plant pathogenic Ralstonia spp. PeerJ 7:e7346. doi: 10.7287/peerj.preprints.27726v1
Safni, I., Cleenwerck, I., De Vos, P., Fegan, M., Sly, L., and Kappler, U. (2014). Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. Int. J. Syst. Evol. Microbiol. 64, 3087–3103. doi: 10.1099/ijs.0.066712-0
Schultink, A., Qi, T., Lee, A., Steinbrenner, A. D., and Staskawicz, B. (2017). Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. 92, 787–795. doi: 10.1111/tpj.13715
Schwartz, A. R., Potnis, N., Timilsina, S., Wilson, M., Patané, J., Martins, J., et al. (2015). Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Front. Microbiol. 6:535.
Sharma, S., and Bhattarai, K. (2019). Progress in developing bacterial spot resistance in tomato. Agronomy 9:26. doi: 10.3390/agronomy9010026
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Sohn, K. H., Segonzac, C., Rallapalli, G., Sarris, P. F., Woo, J. Y., Williams, S. J., et al. (2014). The nuclear immune receptor RPS4 is required for RRS1SLH1-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genet. 10:e1004655. doi: 10.1371/journal.pgen.1004655
Staskawicz, B. J., and Schultink, A. C. (2019). Roq1 Provides Resistance to Both Xanthomonas and Pseudomonas in Plants. World Patent WO2019040483. Geneva: WIPO.
Tai, T. H., Dahlbeck, D., Clark, E. T., Gajiwala, P., Pasion, R., Whalen, M. C., et al. (1999). Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. U.S.A. 96, 14153–14158. doi: 10.1073/pnas.96.24.14153
Teper, D., Salomon, D., Sunitha, S., Kim, J.-G., Mudgett, M. B., and Sessa, G. (2014). Xanthomonas euvesicatoria type III effector XopQ interacts with tomato and pepper 14-3-3 isoforms to suppress effector-triggered immunity. Plant J. 77, 297–309. doi: 10.1111/tpj.12391
Thomas, N. C., Hendrich, C. G., Gill, U. S., Allen, C., Hutton, S. F., and Schultink, A. (2019). Roq1 confers resistance to Xanthomonas, Pseudomonas syringae and Ralstonia solanacearum in tomato. bioRxiv [Preprint]. doi: 10.1101/813758
Vasse, J., Frey, P., and Trigalet, A. (1995). Microscopic Studies of Intercellular Infection and Protoxylem Invasion of Tomato Roots by Pseudomonas Solanacearum. Available at: https://pubag.nal.usda.gov/catalog/1443867 (Accessed July 1, 2019).
Vincelli, P. (2016). Genetic engineering and sustainable crop disease management: opportunities for case-by-case decision-making. Sustain. Sci. Pract. Policy 8:495. doi: 10.3390/su8050495
Wei, C. F., Kvitko, B. H., Shimizu, R., Crabill, E., Alfano, J. R., Lin, N. C., et al. (2007). A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 51, 32–46. doi: 10.1111/j.1365-313x.2007.03126.x
Xin, X.-F., Kvitko, B., and He, S. Y. (2018). Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol. 16, 316–328. doi: 10.1038/nrmicro.2018.17
Yuliar, Nion, Y. A., and Toyota, K. (2015). Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes Environ. 30, 1–11. doi: 10.1264/jsme2.me14144
Keywords: plant immunity, Ralstonia, Xanthomonas, Pseudomonas, tomato
Citation: Thomas NC, Hendrich CG, Gill US, Allen C, Hutton SF and Schultink A (2020) The Immune Receptor Roq1 Confers Resistance to the Bacterial Pathogens Xanthomonas, Pseudomonas syringae, and Ralstonia in Tomato. Front. Plant Sci. 11:463. doi: 10.3389/fpls.2020.00463
Received: 24 December 2019; Accepted: 30 March 2020;
Published: 23 April 2020.
Edited by:
Marc Valls, University of Barcelona, SpainReviewed by:
Nemo Peeters, Institut National de la Recherche Agronomique (INRA), FranceKee Hoon Sohn, Pohang University of Science and Technology, South Korea
Kouhei Ohnishi, Kochi University, Japan
Copyright © 2020 Thomas, Hendrich, Gill, Allen, Hutton and Schultink. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alex Schultink, alex.schultink@fortiphyte.com
 Nicholas C. Thomas1,2
Nicholas C. Thomas1,2 Connor G. Hendrich
Connor G. Hendrich Upinder S. Gill
Upinder S. Gill